6 Bootstrapping
library(PSAboot)
boot.matching.1to3 <- function(Tr, Y, X, X.trans, formu, ...) {
return(boot.matching(Tr=Tr, Y=Y, X=X, X.trans=X.trans, formu=formu, M=3, ...))
}
boot_out <- PSAboot(Tr = lalonde$treat == 1,
Y = lalonde$re78,
X = lalonde[,all.vars(lalonde.formu)[-1]],
seed = 2112,
methods=c('Stratification' = boot.strata,
'ctree' = boot.ctree,
'rpart' = boot.rpart,
'Matching' = boot.matching,
'Matching-1-to-3' = boot.matching.1to3,
'MatchIt' = boot.matchit) )
summary(boot_out)
plot(boot_out)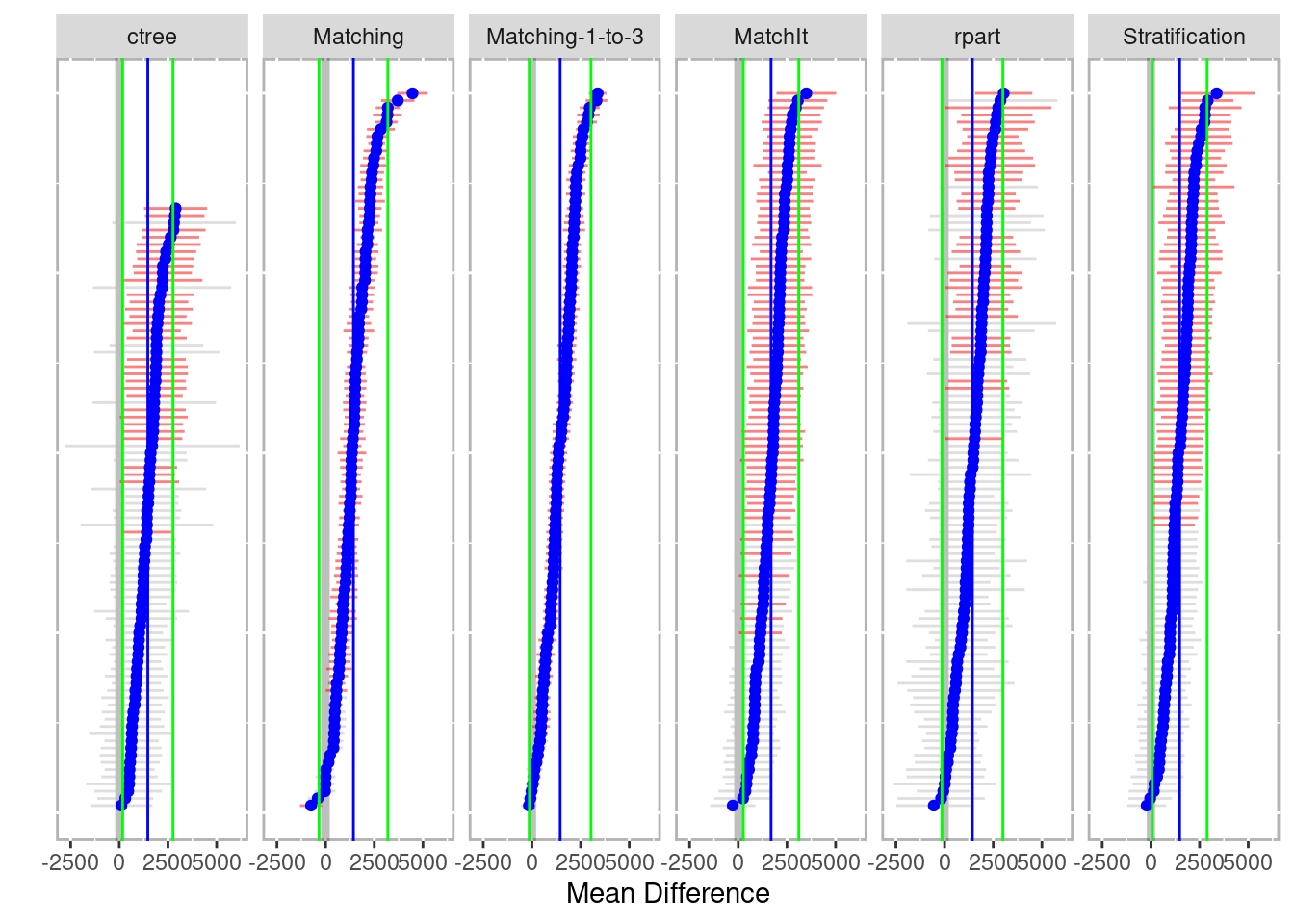
Figure 6.1: Mean difference across all bootstrap samples by method
boxplot(boot_out)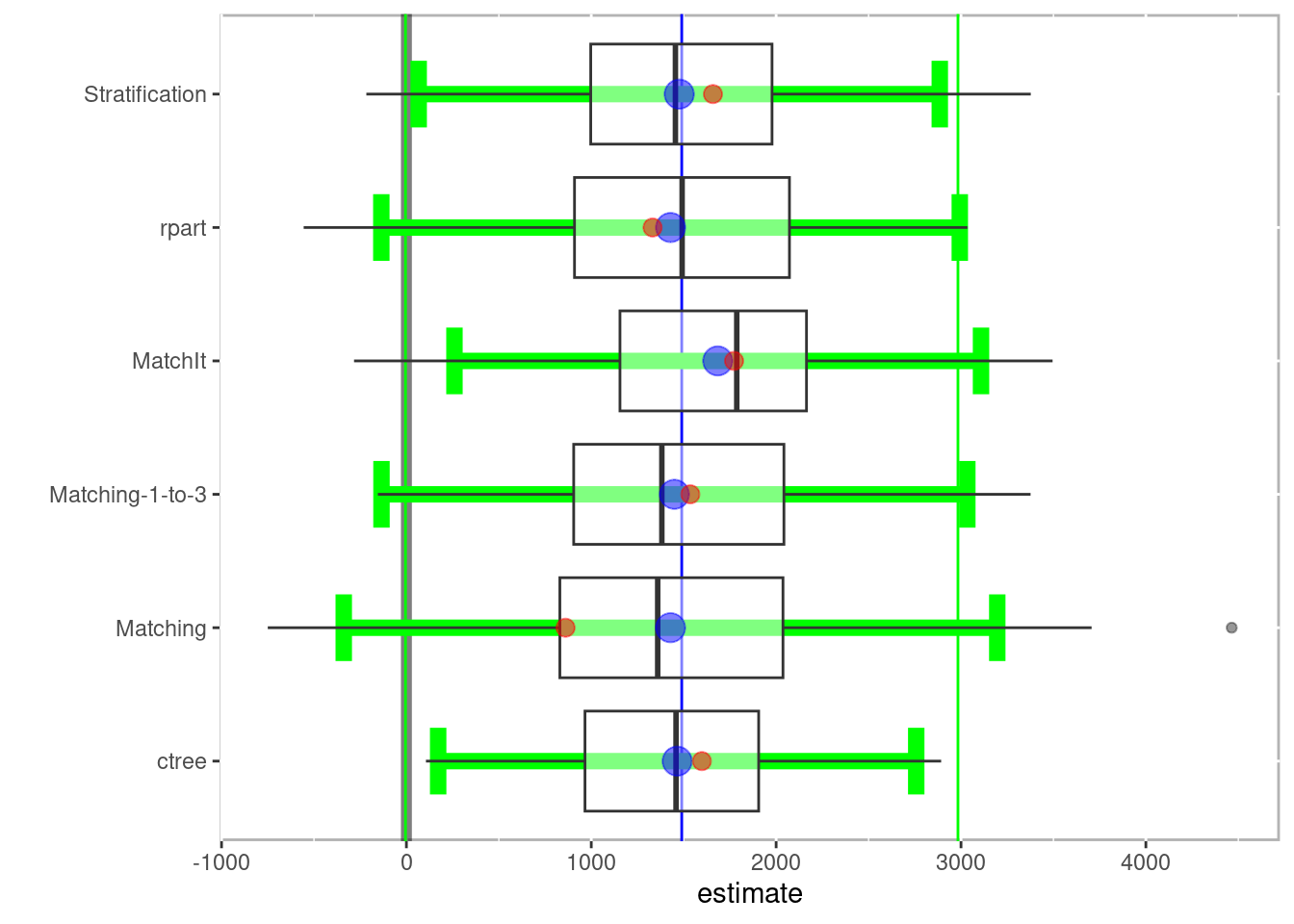
matrixplot(boot_out)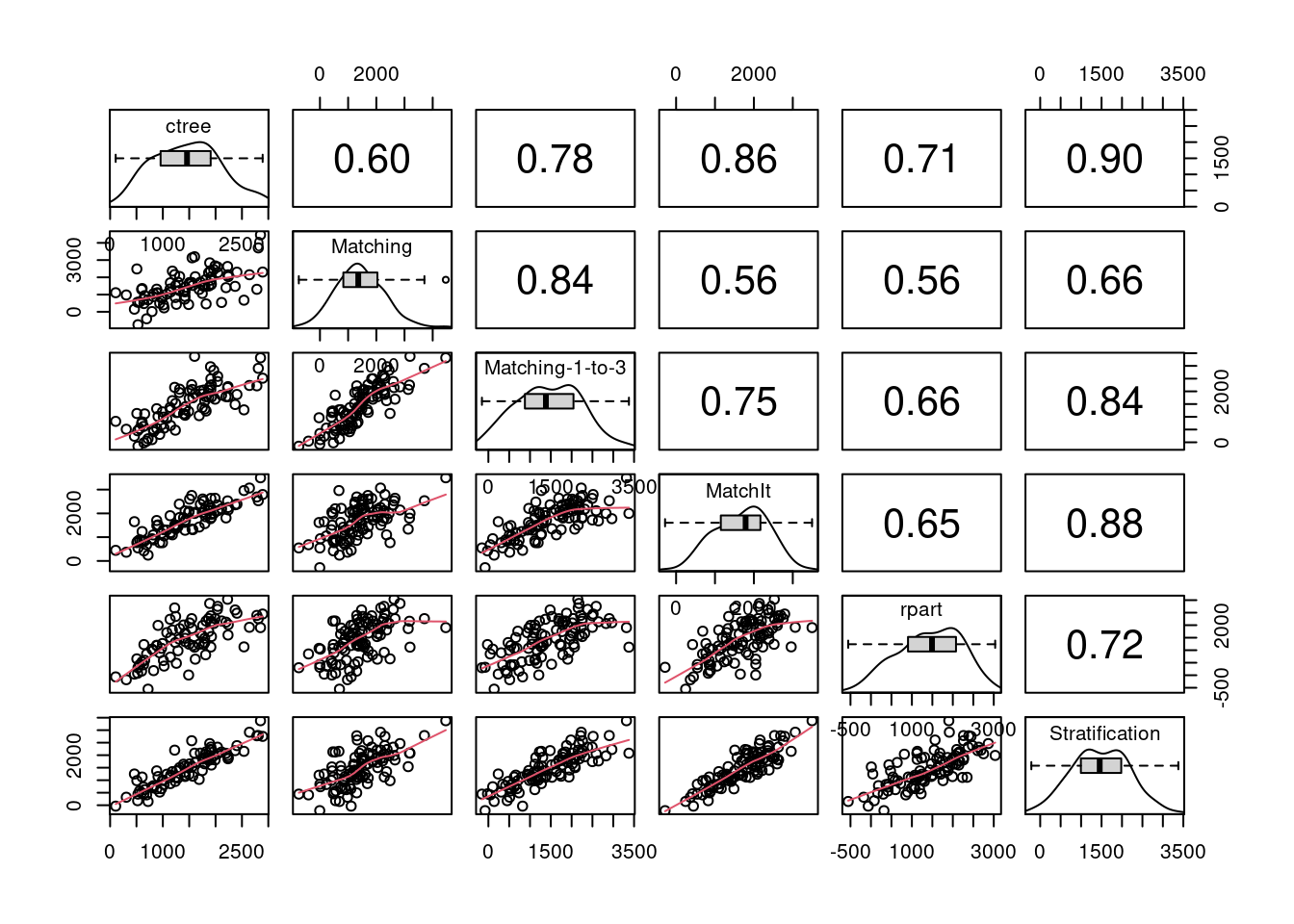
boot_balance <- balance(boot_out)
boot_balance## Unadjusted balance: 1.48309081605193## 0.04
## 0.08
## 0.05
## 0.06
## 0.01
## 0.06
## 0.04
## NA
## 0.07
## 0.07
## 0.05
## 0.08
plot(boot_balance)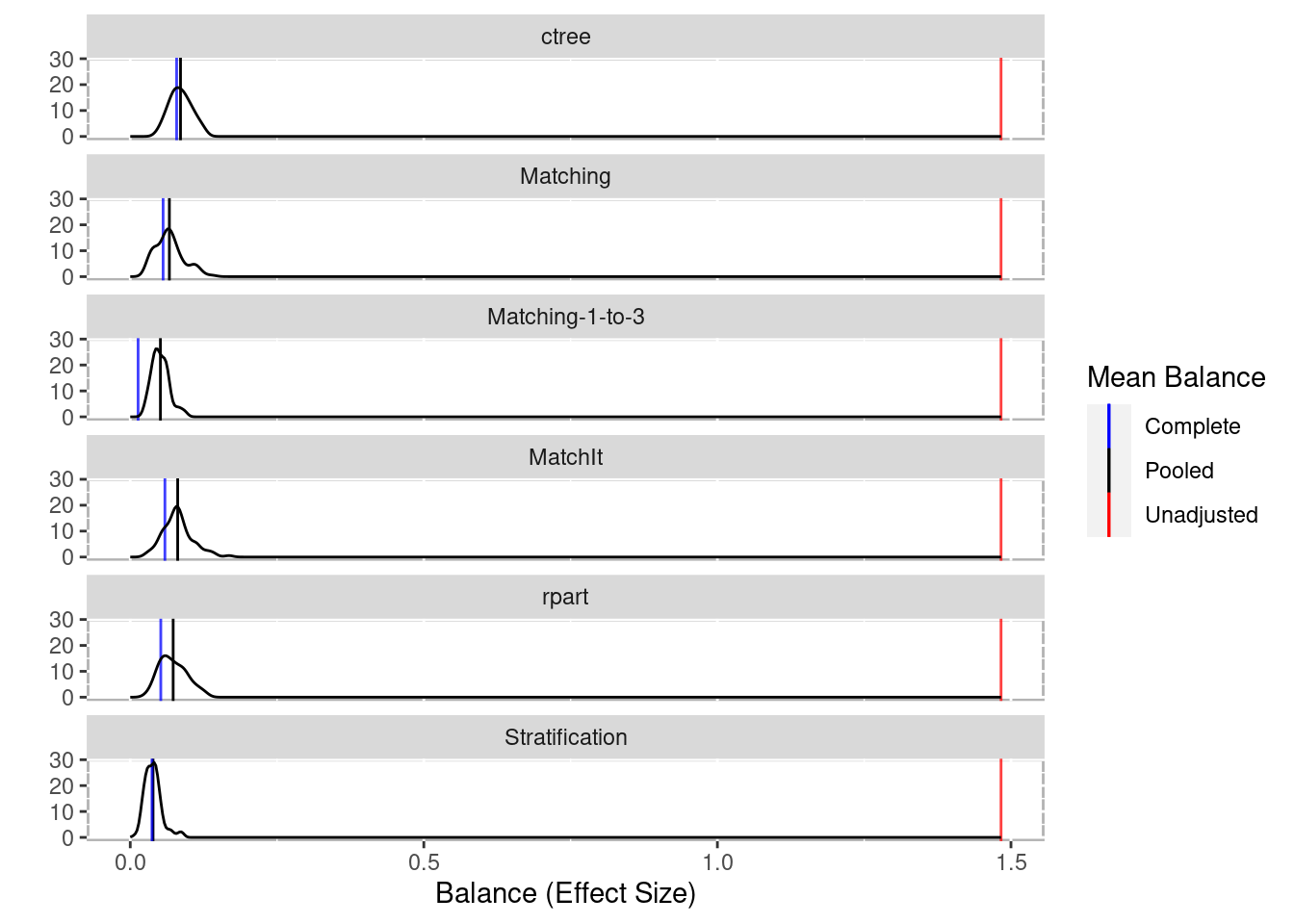
boxplot(boot_balance) + geom_hline(yintercept=.1, color='red')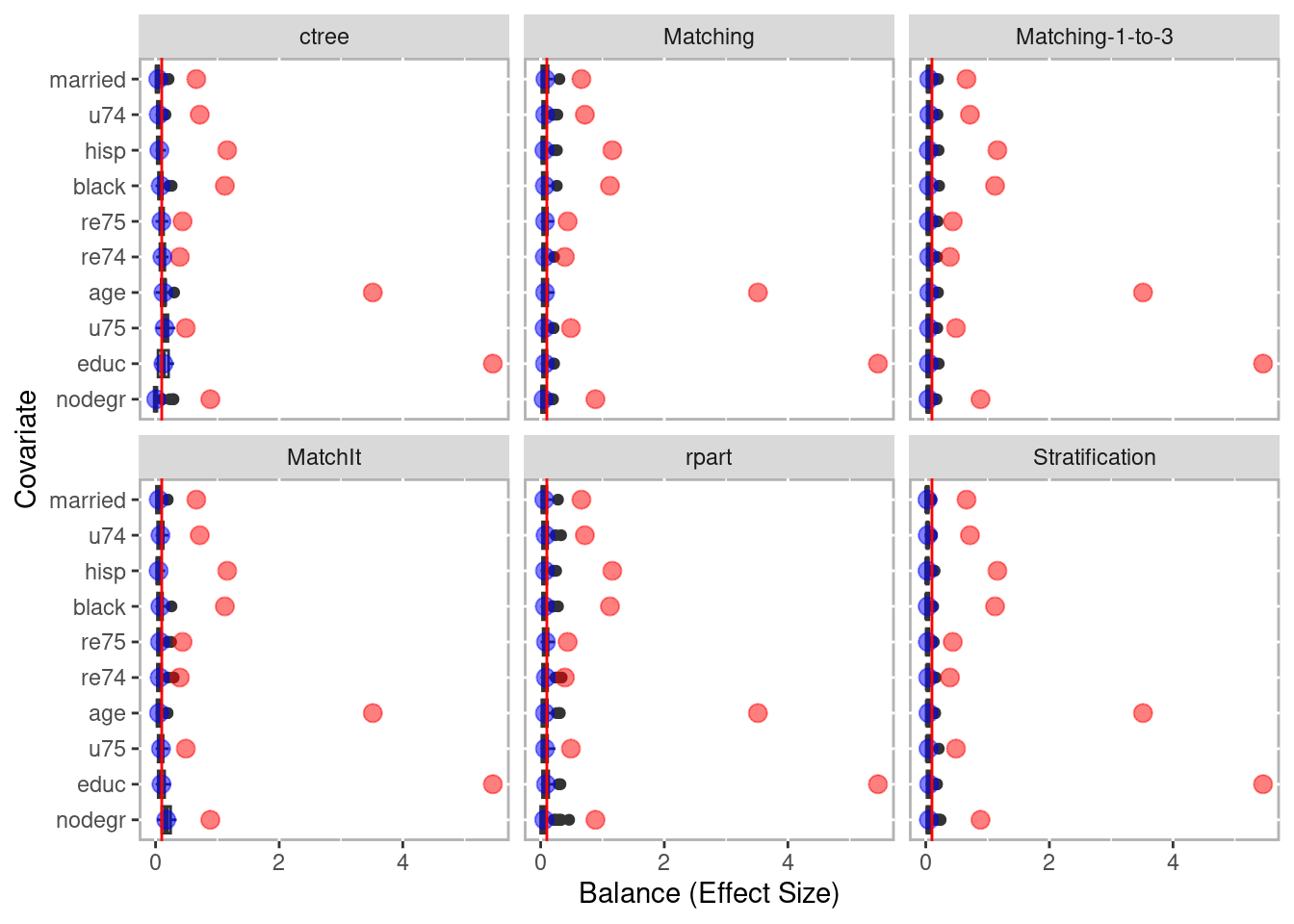
Details are available within the returned object
boot_balance$unadjusted## 3.51
## 5.46
## 1.12
## 1.16
## 0.66
## 0.89
## 0.39
## 0.44
## 0.72
## 0.49
boot_balance$complete## 0.04
## 0.00
## 0.00
## 0.01
## 0.00
## 0.16
## 0.03
## 0.16
## 0.05
## 0.04
## 0.02
## 0.06
## 0.09
## 0.15
## 0.14
## 0.04
## 0.01
## 0.02
## 0.04
## 0.05
## 0.08
## 0.04
## 0.00
## 0.09
## 0.02
## 0.08
## 0.10
## 0.09
## 0.01
## 0.02
## 0.03
## 0.08
## 0.02
## 0.09
## 0.02
## 0.07
## 0.00
## 0.09
## 0.05
## 0.06
## 0.00
## 0.00
## 0.03
## 0.09
## 0.01
## 0.07
## 0.01
## 0.04
## 0.06
## 0.06
## 0.06
## 0.02
## 0.05
## 0.08
## 0.03
## 0.03
## 0.01
## 0.10
## 0.00
## 0.05
boot_balance$pooled |> head()## 0.02
## 0.03
## 0.04
## 0.03
## 0.04
## 0.05
## 0.07
## 0.09
## 0.12
## NA
## 0.06
## 0.12
## 0.05
## 0.09
## 0.08
## 0.11
## 0.08
## 0.07
## 0.06
## 0.07
## 0.03
## 0.07
## 0.05
## 0.06
## 0.04
## 0.03
## 0.03
## 0.08
## 0.03
## 0.05
## 0.08
## 0.10
## 0.09
## 0.07
## 0.10
## 0.10